Understanding PTMs: Insights into Protein Regulation and Functional Implications
In this paper, we will be focusing, by way of example, on ubiquitination among other PTMs, elucidating on their functionalities, current research focuses, and selective investigative strategies.

Since the last century, our understanding of post-translational modifications (PTMs) in proteins has significantly deepened. Over the past decade, further insight has been gained through the discovery of novel protein PTMs, the novel development of whole-genome maps, and the inventive strategies for localized and temporal control of PTM deposition or removal. In this digital age, with the widespread availability of vast amounts of data from various biological systems, the functional roles of PTMs are gradually being revealed in key processes such as transcription, recombination, replication, DNA repair, and genomic structural regulation.
With a broad array of types, post-translational modifications exhibit unique features specific to various proteins. Methylation is the most common form of modification that influences protein structure and interactions with other proteins, implicated in virtually all biological processes. Phosphorylation, on the other hand, can alter protein conformation, activating or inhibiting catalytic activity. Recently, novel areas of research and emerging trends are becoming more noticeable involving other forms of modifications. In this paper, we will be focusing, by way of example, on ubiquitination among other PTMs, elucidating on their functionalities, current research focuses, and selective investigative strategies.
Acetylation
Acetylation impacts protein conformation and affinity with other proteins, participating in the regulation of transcription and metabolism.
Nε Acetylation, colloquially termed "acetylation", is a pervasive and reversible PTM, orchestrated by two classes of enzymes: Lysine acetyltransferases (KATs) and Lysine deacetylases (KDACs). KATs primarily catalyze the transfer of acetyl groups onto Lys residues, while KDACs facilitate the removal of acetyl groups. The KDACs are subdivided into two subgroups: the Zn2+ dependent histone deacetylases (HDACs) and the NAD+-dependent sirtuins. More than 40 different Lys residues in the four canonical core histones H2A, H2B, H3, and H4 can be acetylated. Additionally, proteomic studies have revealed acetylation phenomena in thousands of non-histone proteins in the cell, affecting processes such as metabolism, RNA processing, translation, protein folding, chromatin organization, protein degradation, and cytoskeleton organization. Recent studies also view acetylation as a means to identify candidate disease biomarkers and have found that histone acetylation can mediate cellular memory. This capacity for acetylation permits the coupling of cell state with transcriptional output and cell fate decisions.
PTMs like phosphorylation, ubiquitination, and Acetylation play significant roles in autophagy regulation, with acetylation being a pivotal regulatory mechanism of autophagy. Acetylation influences autophagy initiation and autophagosome formation by targeted regulation of core components like ULK1 complex, BECN1-PIK3C3 complex, and LC3 lipidation system. Further, recent research suggests that acetylation occurs on key proteins involved in autophagic assembly and autophagosome-lysosome fusion, such as SQSTM1/p62 and STX17. Additionally, acetylation also regulates autophagy at the transcriptional level by targeting histones and the transcription factor TFEB.
Ubiquitination: An Essential Process in Protein Degradation
Ubiquitination, the covalent linkage of ubiquitin to proteins, is a prevalent post-translational modification (PTM) with significant implications for the stability, subcellular positioning, functionality, and complex formation of substrate proteins. The ubiquitination process is catalyzed by a collection of enzymatic reactions involving ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin-ligating enzymes (E3). In the final step, an isopeptide bond forms between the carboxyl group of the terminal glycine residue on ubiquitin and the epsilon amino group of a lysine residue in the substrate protein.
The ubiquitin superfamily is a tremendous set of proteins, also involving autophagy-related (ATG) 8, ATG12, neural precursor cell expressed developmentally down-regulated protein 8 (NEDD8), small ubiquitin-related modifier (SUMO), human leukocyte antigen F adjacent transcript 10 (FAT10), among others. This superfamily also includes the smaller subfamilies of interferon-stimulated gene 15 kDa (ISG15) and ubiquitin-fold modifier 1 (UFM1). Like ubiquitin, these family proteins also establish covalent links with their substrates through analogous ubiquitin system reactions.
Recent studies have directed attention to the formation of ester bonds between ubiquitin's serine or threonine residues (rather than the terminal glycine) and phospholipids, while the exact connection site remains to be identified. A recent investigation has revealed that mammalian cells' host E3 ligase, the RING finger (RNF) 213, ubiquitinates Salmonella lipopolysaccharide, facilitating bacterial removal via autophagy. In this case, ubiquitin may form an ester bond with lipopolysaccharides instead of a typical peptide bond, but the precise linkage mode remains undetermined.
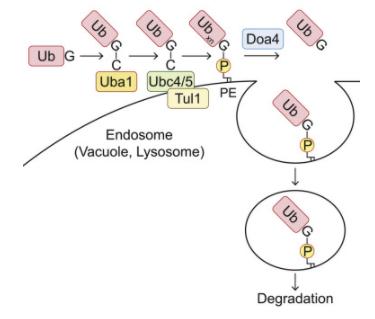
Within the ubiquitin family, the ATG8 subfamily uniquely features a prominent association with phospholipids. Existing research has unveiled that ubiquitin forms bonds with phospholipids, notably phosphatidylethanolamine (PE), in both yeast and mammalian cells. To investigate whether ubiquitin interacts with organelle membrane phospholipids, it is feasible to purify ubiquitin species possessing hydrophobic traits from samples. For instance, solubilizing the cell membrane from cells expressing 3×FLAG-ubiquitin in a buffer containing 1% TritonX-114, followed by heating to the cloud point to achieve phase partitioning. Ubiquitin can then be immunoprecipitated from the TritonX-114 fraction using an anti-FLAG antibody. In addition to a substantial amount of polyubiquitin species, a band with a higher molecular weight (Ub*) can also be detected in the hydrophobic sector of TritonX-114. After treatment with phospholipase D (PLD), which hydrolyzes the diester bond of glycerophospholipids, this high molecular weight entity shifts downwards, thereby confirming the association of ubiquitin with phospholipids.
To detect ubiquitinated phospholipids at endogenous levels within cells, hydrophobic ubiquitin species can be recovered from yeast membranes through affinity purification using ubiquitin-related (UBA) domain of ubiquilin 1 (UQ1). Diand triubiquitin conjugates, which can be cleaved by PLD, can be detected in the TritonX-114 detergent phase. The absence of ubiquitin monomers in the TritonX-114 phase could be attributed to the attributes of UQ1UBA, as it tends to capture oligo- and polyubiquitin rather than monoubiquitin. Moreover, the multiphospholipidation status of endogenous phospholipids can be studied via lipid extraction assays.
To determine the types of phospholipids that bind to ubiquitin in the sample, phosphatidylserine (PS) and PE can be considered, as they are potential ubiquitination targets, having an amino group capable of forming an amide bond with the C-terminal glycine of ubiquitin and known to bind with ATG8 family proteins. Subsequently, gel-internal saponification (acyl chain cleavage), leucine aminopeptidase digestion, and tandem mass spectrometry (MS/MS) analysis can be performed on Ub*.
To ascertain the location of Ub-PE, cell membranes can be isolated via membrane flotation in discontinuous iodixanol gradients, followed by TritonX-114 phase partitioning. When further identifying the enzyme catalyzing PE ubiquitination, the auxin-induced degron system can be adopted to deplete ubiquitin-activating enzyme (E1) Uba1, to inhibit PE ubiquitination. To confirm that the identified enzyme catalyzes the sufficient transfer of ubiquitin to PE, the process can be reconstituted in vitro using liposomes composed of 40 mol% PE or PS and 50 mol% phosphatidylcholine (PC).
For a more thorough investigation of whether the hydrophobic ubiquitin species detected in the cell indeed contain Ub-PE, Ub-PE reconstituted in vitro can serve as a molecular standard. By using MS/MS spectra of C-terminal peptides with or without GPE modifications derived from in vitro reconstituted Ub-PE (post-saponification and leucine aminopeptidase treatment), peptide quantification based on mass spectrometry can be established through a combination of selective reaction monitoring (SRM) method and stable isotope labeling. Subsequently, further investigations can then explore whether other ubiquitin-like proteins are also binding to phospholipids, thereby expanding on the existing findings.
Sumoylation: Subcellular Localization and Protein Interactions
Sumoylation, a modification involving small ubiquitin-related modifiers (SUMOs), is an emerging form of ubiquitin-like molecular modification. Like ubiquitination, it is integral to crucial post-translational modifications (PTMs). Sumoylation events are catalyzed by a minor modification enzyme, dynamically regulating a multitude of target proteins. SUMOs can attach to target proteins either as individual monomers or in various types of polymeric forms. Non-covalent receptors recognize Sumoylated proteins via SUMO interaction motifs.
Sumoylation acts upon functionally partnered proteins to control core nuclear processes, encompassing gene expression, the DNA damage response, RNA processing, the progress of the cell cycle, and protein stability. Recent research advances have deepened our understanding of the cellular and pathophysiological roles of Sumoylation, extending its function to the regulation of immunity, pluripotency, and nucleolar assembly in response to oxidative stress, partly realized through the recently characterized liquid-liquid phase separation mechanism.
The breakthrough in understanding the action and regulation of Sumoylation has paved new paths for disease treatment targeting Sumoylation. The first Sumoylation inhibitor is currently in clinical trials and is being explored as a potential anti-cancer drug.
What's Your Reaction?




















